import pandas as pd
import statsmodels as sm
import statsmodels.formula.api as smf
import pymc as pm
import numpy as np
Ordinary least squares using PyMC#
Recall the ordinary least squares model you might be familiar with from a previous statistics or econometrics class. We are interested in estimating the parameter vector \(\beta\) from the following population regression function:
where \(\mathbf{y}\) and \(\mathbf{\epsilon}\) are each \(N \times 1\) vectors of independent (outcome) and unobservable random variables. The \(N \times K\) matrix \(\mathbf{x}\) are the non-random (assumed for this notebook) independent variables (sometimes referred to as covariates). The least squares approach yields the “ols” estimator
The frequentist approach performs inference based on the variance covariance matrix of \(\mathbf{b}\) which is estimated as
where \(\hat{\sigma}^2 = \frac{(\mathbf{y-xb})'(\mathbf{y-xb})}{N-K}\)
The square root of the diagonal of this matrix yields standard errors for estimated coefficients as reported in programs like Stata. These standard errors are used for inference related questions.
The Bayesian approach to OLS takes a different approach. Consider the model
We can setup the likelihood and priors for getting information about the posterior. Assembling the pieces,
Likelihood:
\[ prob(\mathbf{y}| \mathbf{x}, \mathbf{\beta}, \mathbf{\sigma}) = \prod_{i=1}^N \frac{1}{\sqrt{2\pi\sigma^2}} e^\frac{(y_i-\mathbf{x}_i \mathbf{b})^2}{2\sigma^2} \]Prior on \(\sigma\):
\[ prob(\sigma | a=0.01, b=10) = \frac{1}{(10 - .01)} \text{ for } .01 \le \sigma \le 10 \text{, 0 otherwise} \]Prior on \(\mathbf{\beta}\)
\[ prob(\mathbf{b} | \mu_b = 0, \sigma_b = 10) = \prod_{k = 1}^{K} \frac{1}{\sqrt{2\pi \sigma^2_b}} e ^ {\frac{b_k - \mu_b}{2\sigma^2_b}} \]
Let \(\theta\) be the vector of all unknown parameters, \(\begin{bmatrix} \beta_1 \\ \vdots \\ \beta_k \\ \vdots \\ \beta_K \\ \sigma \end{bmatrix}\). Then the posterior is proportional to
An example#
We will use the Tobias and Koop dataset for this excercise. We’ll estimate a simple model using frequentist and Bayesian methods. This dataset has the following fields:
Variable |
Definition |
|---|---|
id |
person id, ranging from 1 to 2,178 |
educ |
education (years) |
ln_wage |
log of hourly wage |
pexp |
potential experience |
time |
time trend |
ability |
ability |
meduc |
mother’s education (years) |
feduc |
father’s education |
broken_home |
dummy variable for residence in a broken home |
siblings |
number of siblings |
pexp2 |
squared potential experience |
tobias_koop = pd.read_stata('https://rlhick.people.wm.edu/econ407/data/tobias_koop.dta')
tobias_koop.head()
| id | educ | ln_wage | pexp | time | ability | meduc | feduc | broken_home | siblings | pexp2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.0 | 13.0 | 1.82 | 1.0 | 0.0 | 1.0 | 12.0 | 12.0 | 0.0 | 1.0 | 1.0 |
| 1 | 1.0 | 18.0 | 3.29 | 3.0 | 7.0 | 1.0 | 12.0 | 12.0 | 0.0 | 1.0 | 9.0 |
| 2 | 1.0 | 18.0 | 3.21 | 5.0 | 9.0 | 1.0 | 12.0 | 12.0 | 0.0 | 1.0 | 25.0 |
| 3 | 1.0 | 18.0 | 3.06 | 6.0 | 10.0 | 1.0 | 12.0 | 12.0 | 0.0 | 1.0 | 36.0 |
| 4 | 2.0 | 15.0 | 2.14 | 4.0 | 6.0 | 1.5 | 12.0 | 12.0 | 0.0 | 1.0 | 16.0 |
Note, that this is a panel dataset having the following time periods:
tobias_koop.time.value_counts()
time
12.0 1570
11.0 1535
10.0 1520
13.0 1517
14.0 1499
9.0 1443
8.0 1398
7.0 1284
6.0 1248
5.0 1093
4.0 1034
3.0 964
2.0 745
1.0 615
0.0 454
Name: count, dtype: int64
For purposes of this lecture we will ignore the panel aspects of this
dataset and estimate an ordinary least squares model. Furthermore,
due to memory constraints that students may encounter on jupyterhub
for some of the features below, we will restrict our sample to
time==4 (1983). In general we don’t recommend omitting data like
this, but do it here for the aforementioned reasons. Reducing the
data:
print("Records in original dataset: ", tobias_koop.shape[0])
tobias_koop = tobias_koop[tobias_koop.time == 4]
print("Records in reduced dataset: ", tobias_koop.shape[0])
Records in original dataset: 17919
Records in reduced dataset: 1034
Let’s estimate a simple wage model:
Using statsmodels, we have
res = smf.ols(formula='ln_wage ~ educ + pexp + broken_home',
data=tobias_koop).fit()
print(res.summary())
OLS Regression Results
==============================================================================
Dep. Variable: ln_wage R-squared: 0.142
Model: OLS Adj. R-squared: 0.140
Method: Least Squares F-statistic: 57.04
Date: Fri, 12 Jan 2024 Prob (F-statistic): 4.09e-34
Time: 10:52:15 Log-Likelihood: -598.31
No. Observations: 1034 AIC: 1205.
Df Residuals: 1030 BIC: 1224.
Df Model: 3
Covariance Type: nonrobust
===============================================================================
coef std err t P>|t| [0.025 0.975]
-------------------------------------------------------------------------------
Intercept 0.6789 0.133 5.101 0.000 0.418 0.940
educ 0.0874 0.009 9.290 0.000 0.069 0.106
pexp 0.0804 0.007 12.034 0.000 0.067 0.093
broken_home -0.0031 0.036 -0.085 0.933 -0.074 0.068
==============================================================================
Omnibus: 49.553 Durbin-Watson: 1.734
Prob(Omnibus): 0.000 Jarque-Bera (JB): 95.369
Skew: -0.327 Prob(JB): 1.95e-21
Kurtosis: 4.336 Cond. No. 132.
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
Bayesian Estimation#
We will continue using the model setup as specified above,
The implementation in PyMC in code is very close to this model definition:
with pm.Model() as ols_model:
# priors
b0 = pm.Normal('constant', mu=0., sigma=10.)
b1 = pm.Normal('educ', mu=0., sigma=10.)
b2 = pm.Normal('pexp', mu=0., sigma=10.)
b3 = pm.Normal('broken_home', mu = 0., sigma=10.)
sigma = pm.Uniform('sigma', lower=.1, upper=10.)
# yhat (or the mean for each i's likelihood contrib)
mu_i = b0 + b1 * tobias_koop.educ + b2 * tobias_koop.pexp +\
b3 * tobias_koop.broken_home
# likelihood
like = pm.Normal('likelihood', mu=mu_i, sigma=sigma,
observed = tobias_koop.ln_wage)
Note, that MAP (maximum of the posterior) is very close to the OLS estimate:
with ols_model:
startvals = pm.find_MAP()
print(startvals)
{'constant': array(0.67877072), 'educ': array(0.0874166), 'pexp': array(0.08036808), 'broken_home': array(-0.00305453), 'sigma_interval__': array(-3.36234812), 'sigma': array(0.43158197)}
Note
On our jupyterhub server, it is necessary to include two function arguments, which in normal cases you might not want to use:
Since each user is allocated 2 CPU cores. For PyMC to run properly, you must use the
cores=2argument below. While the code will run without this argument, results may be unreliable particularly for this notebook. On a typical PC, you would want to omit thecoresargument and let PyMC use the maximum number of cores available for quickest execution.The
progressbar=Falseis included so that these notes will look better on this webpage. In general, you would probably want to view the progress and wouldn’t need this option.
Sampling from the posterior for the model is accomplished using
with ols_model:
step = pm.Metropolis()
samples = pm.sample(1000, step=step, tune=500, initvals=startvals, cores=2, chains=4,
return_inferencedata=True, progressbar=False)
Multiprocess sampling (4 chains in 2 jobs)
CompoundStep
>Metropolis: [constant]
>Metropolis: [educ]
>Metropolis: [pexp]
>Metropolis: [broken_home]
>Metropolis: [sigma]
Sampling 4 chains for 500 tune and 1_000 draw iterations (2_000 + 4_000 draws total) took 2 seconds.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for details
Summarizing the trace:
pm.summary(samples)
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| constant | 0.751 | 0.081 | 0.649 | 0.932 | 0.035 | 0.026 | 6.0 | 15.0 | 1.85 |
| educ | 0.083 | 0.006 | 0.072 | 0.093 | 0.002 | 0.002 | 7.0 | 14.0 | 1.70 |
| pexp | 0.077 | 0.006 | 0.066 | 0.087 | 0.002 | 0.001 | 13.0 | 87.0 | 1.24 |
| broken_home | -0.007 | 0.035 | -0.080 | 0.051 | 0.003 | 0.002 | 169.0 | 323.0 | 1.04 |
| sigma | 0.432 | 0.010 | 0.414 | 0.449 | 0.001 | 0.000 | 241.0 | 186.0 | 1.03 |
The summary above shows
Results very similar to OLS
That we have very doubtful chain convergence as the Gelman-Rubin score (
r_hatis large)
Examining autocorrelation, we have
pm.plot_autocorr(samples, combined=True);
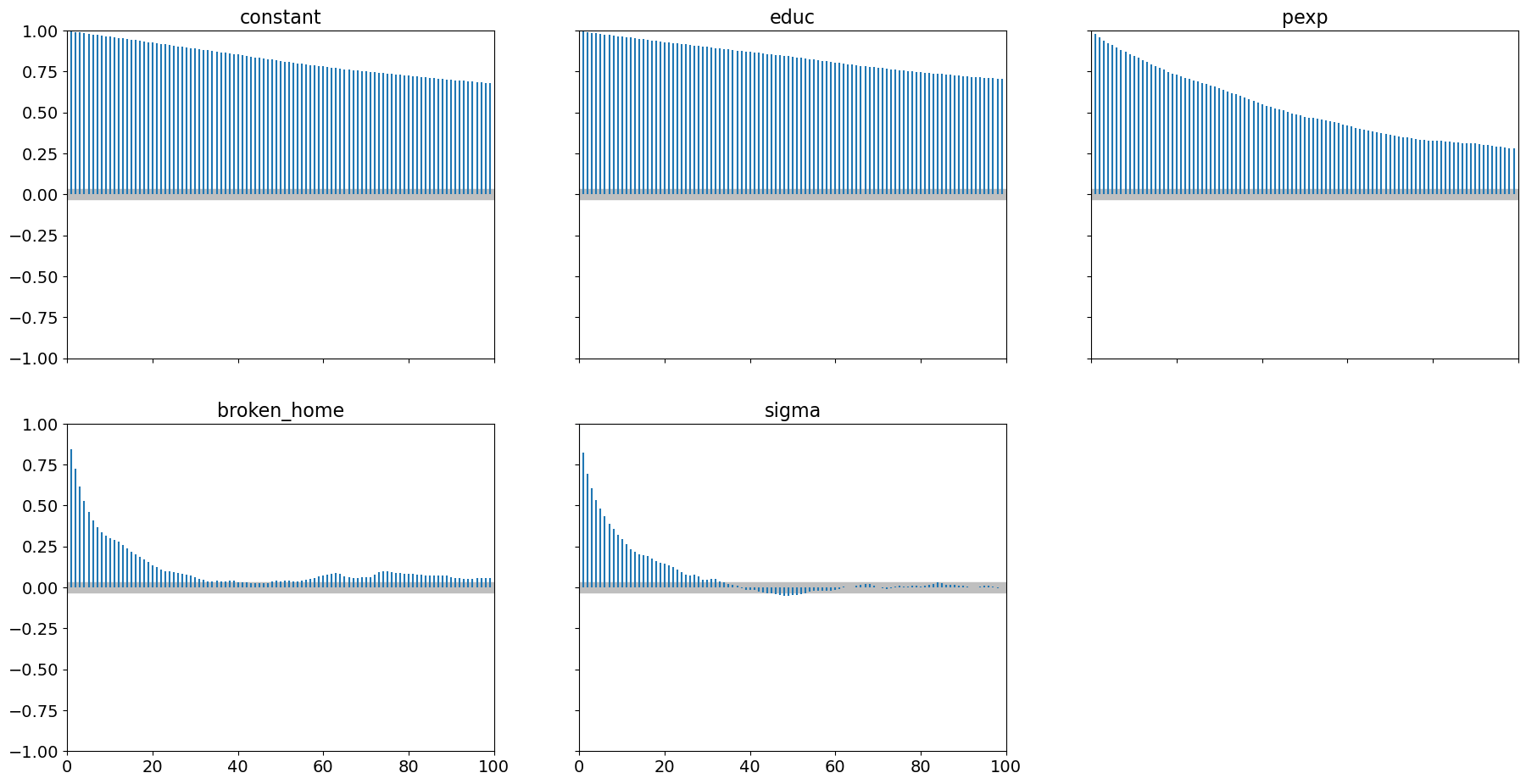
which shows very high level of autocorrelation particularly for educ
and the model constant.
The accept rate is
accept = samples.sample_stats.accepted.mean().to_numpy()
print("Acceptance Rate: ", accept.round(3))
Acceptance Rate: 0.126
To obtain better results, we need to either
Increase sample size (and tuning), or
Choose a different step method
At this point in the course, our only option is to increase sampling and tuning:
with ols_model:
step = pm.Metropolis()
samples = pm.sample(10000, step=step, tune=10000, initvals=startvals, cores=2, chains=4,
return_inferencedata=True, progressbar=False)
Multiprocess sampling (4 chains in 2 jobs)
CompoundStep
>Metropolis: [constant]
>Metropolis: [educ]
>Metropolis: [pexp]
>Metropolis: [broken_home]
>Metropolis: [sigma]
Sampling 4 chains for 10_000 tune and 10_000 draw iterations (40_000 + 40_000 draws total) took 32 seconds.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for details
We get better, albeit not acceptable, convergence diagnostics:
pm.summary(samples)
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| constant | 0.662 | 0.124 | 0.432 | 0.900 | 0.015 | 0.010 | 73.0 | 156.0 | 1.05 |
| educ | 0.089 | 0.009 | 0.072 | 0.105 | 0.001 | 0.001 | 76.0 | 162.0 | 1.05 |
| pexp | 0.081 | 0.007 | 0.068 | 0.093 | 0.000 | 0.000 | 240.0 | 910.0 | 1.02 |
| broken_home | -0.002 | 0.036 | -0.069 | 0.067 | 0.001 | 0.000 | 4752.0 | 7001.0 | 1.00 |
| sigma | 0.433 | 0.009 | 0.415 | 0.451 | 0.000 | 0.000 | 7234.0 | 8344.0 | 1.00 |
but we still have high levels of autocorrelation:
pm.plot_autocorr(samples, combined=True);
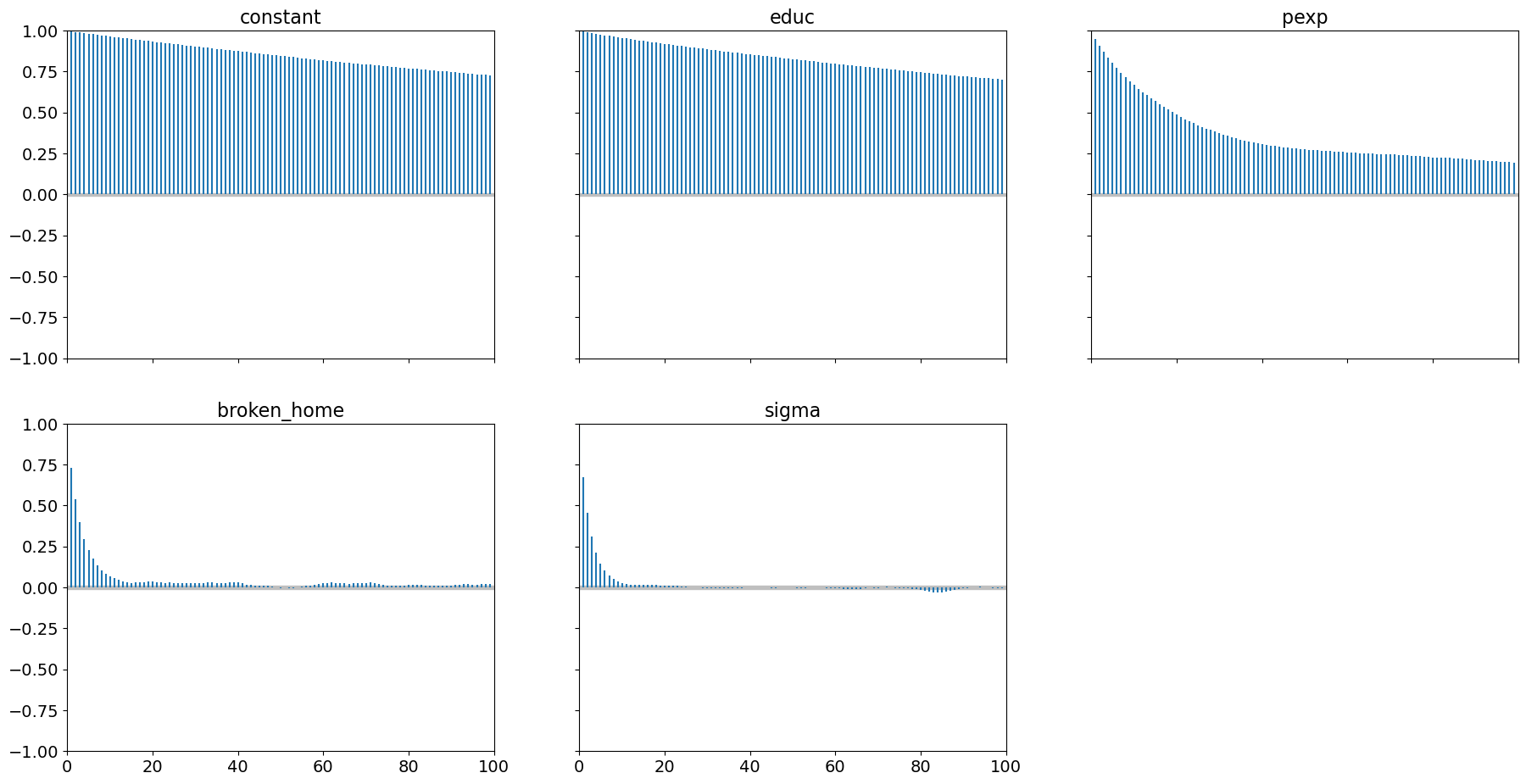
Our acceptance rate is also improved:
accept = samples.sample_stats.accepted.mean().to_numpy()
print("Acceptance Rate: ", accept.round(3))
Acceptance Rate: 0.317
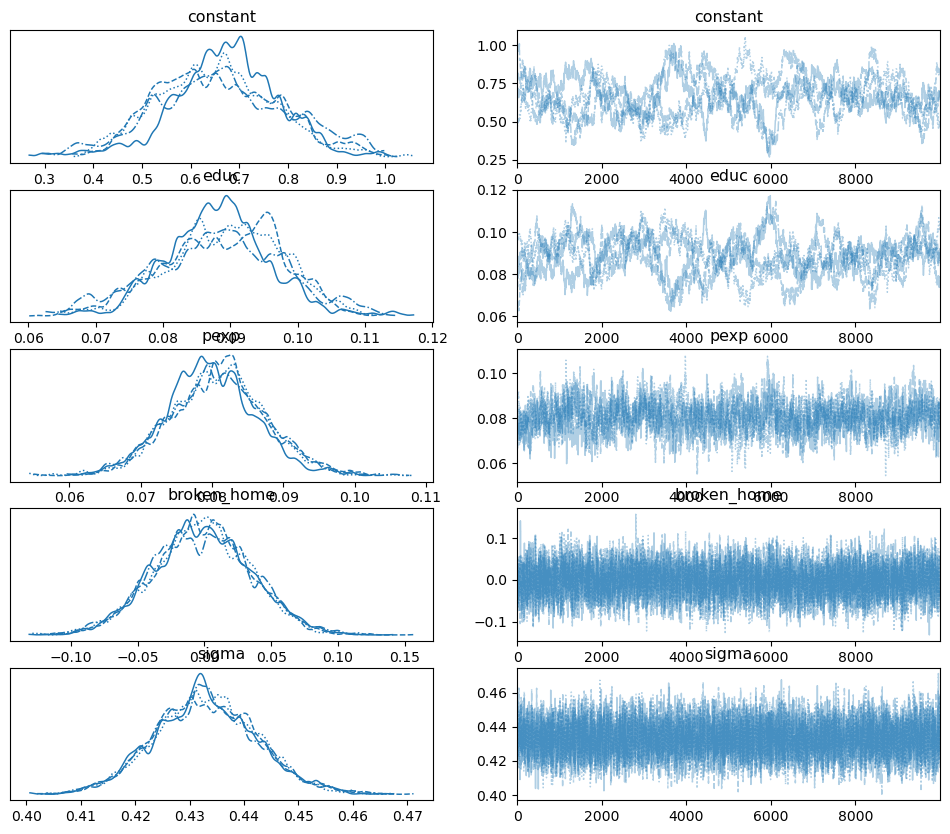
Here is the trace plot:
pm.plot_trace(samples);
Adding “policy relevant” measures to your model#
It is possible to add policy relevant measures in your model and sample from those posteriors quite easily. For demonstration purposes, suppose we want to sample (1) the elasticity of education, and (2) the predicted wage. This is fairly straightforward but we need to redefine our PyMC model to include these items:
Recall that given the \(t^{th}\) sample for \(\beta_{kt}\) in our trace the elasticity in a semi-log model such as ours for individual \(i\) and variable \(k\) is
The average elasticity for variable \(k\) and and sample \(t\) is
The expected elasticity of variable \(k\) as reflected by the underlying uncertainty in our posterior is
where \(\theta\) is the parameter vector and \(p(\theta|x_i, h^0)\) is our posterior and \(h^0\) are hyperparameters describing the priors.
We can approximate this integral using samples from the posterior. Since we have numerous samples for \(b_k\), we estimate the integral above using
where \(T\) is the number of samples in our trace.
To implement this in code, we will ask PyMC to store these
calculations in our sampled chain which we can average to produce the
desired measure. Note the use of pytensor.tensor for performing these
calculations and the syntax pm.Deterministic tells pymc to
include the calculated values in the trace.
import pytensor.tensor as tt
with pm.Model() as ols_model:
## priors
b0 = pm.Normal('constant', mu=0., sigma=10.)
b1 = pm.Normal('educ', mu=0., sigma=10.)
b2 = pm.Normal('pexp', mu=0., sigma=10.)
b3 = pm.Normal('broken_home', mu=0., sigma=10.)
sigma = pm.Uniform('sigma', lower=.1, upper=10.)
## yhat (or the mean for each i's likelihood contrib)
mu_i = b0 + b1 * tobias_koop.educ + b2 * tobias_koop.pexp +\
b3 * tobias_koop.broken_home
## include some policy relevant measures in our samples
# for predicted wage, note the model is in log wage units
yhat = pm.Deterministic('wage_hat', tt.exp(mu_i))
# this is E_{ikt} for a semi log model for each
# sample from our posterior (N x T):
elas_educ = b1 * tobias_koop.educ
# note this will be calculated for **each** observation
# for **each** sampled parameter vector. What we want is
# the average over all observations for each sampled parameter
# vector (E_{kt}) whick we can calculate as
elas = pm.Deterministic('elas_educ', tt.mean(elas_educ, axis=-1))
# likelihood
like = pm.Normal('likelihood', mu=mu_i, sigma=sigma,
observed=tobias_koop.ln_wage)
Now let’s sample from our posterior with these additional elements added in:
with ols_model:
step = pm.Metropolis()
samples = pm.sample(10000, step=step, tune=10000, initvals=startvals, cores=2, chains=4,
return_inferencedata=True, progressbar=False)
Multiprocess sampling (4 chains in 2 jobs)
CompoundStep
>Metropolis: [constant]
>Metropolis: [educ]
>Metropolis: [pexp]
>Metropolis: [broken_home]
>Metropolis: [sigma]
Sampling 4 chains for 10_000 tune and 10_000 draw iterations (40_000 + 40_000 draws total) took 33 seconds.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for details
Note
On some computers, this may not run due to memory limitations or could take a long time to run. If it fails to run on your machine, lower 10000 to some smaller value.
If we examine the samples object, note we have additional items:
samples.posterior
<xarray.Dataset>
Dimensions: (chain: 4, draw: 10000, wage_hat_dim_0: 1034)
Coordinates:
* chain (chain) int64 0 1 2 3
* draw (draw) int64 0 1 2 3 4 5 6 ... 9994 9995 9996 9997 9998 9999
* wage_hat_dim_0 (wage_hat_dim_0) int64 0 1 2 3 4 ... 1030 1031 1032 1033
Data variables:
constant (chain, draw) float64 0.7862 0.7862 0.7919 ... 0.7091 0.7022
educ (chain, draw) float64 0.08177 0.08177 ... 0.08651 0.08651
pexp (chain, draw) float64 0.07368 0.07368 ... 0.07455 0.07455
broken_home (chain, draw) float64 -0.05324 -0.05324 ... 0.04271 0.04271
sigma (chain, draw) float64 0.4054 0.4526 0.4353 ... 0.4195 0.4195
wage_hat (chain, draw, wage_hat_dim_0) float64 6.433 10.05 ... 7.409
elas_educ (chain, draw) float64 1.004 1.004 1.004 ... 1.062 1.062
Attributes:
created_at: 2024-01-12T10:54:02.394867
arviz_version: 0.17.0
inference_library: pymc
inference_library_version: 5.10.3
sampling_time: 33.444486141204834
tuning_steps: 10000In particular, note that we have a new dimension (or coordinate) in
our chain, wage_hat_dim_0 which tracks observations in our data.
Note, predicted wage is a large matrix:
samples.posterior.wage_hat.to_numpy().shape
(4, 10000, 1034)
Since we only have one elasticity measure per sampled parameter vector, it is
samples.posterior.elas_educ.to_numpy().shape
(4, 10000)
in a similar manner to our other parameters:
samples.posterior.constant.to_numpy().shape
(4, 10000)
In our calculations, we elected not to include individual-specific elasticities in our samples, hence there is no dimension over the observations in our data.
Analyzing the Trace#
Summarizing the trace (and omitting our predicted wage) yields:
pm.summary(samples, var_names = ['constant', 'educ', 'pexp', 'broken_home', 'sigma', 'elas_educ'])
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| constant | 0.664 | 0.139 | 0.393 | 0.925 | 0.020 | 0.014 | 51.0 | 53.0 | 1.08 |
| educ | 0.088 | 0.010 | 0.070 | 0.107 | 0.001 | 0.001 | 52.0 | 59.0 | 1.07 |
| pexp | 0.081 | 0.007 | 0.069 | 0.093 | 0.001 | 0.000 | 169.0 | 751.0 | 1.02 |
| broken_home | -0.003 | 0.036 | -0.071 | 0.065 | 0.001 | 0.000 | 3076.0 | 7115.0 | 1.00 |
| sigma | 0.433 | 0.010 | 0.416 | 0.452 | 0.000 | 0.000 | 7719.0 | 8559.0 | 1.00 |
| elas_educ | 1.085 | 0.121 | 0.854 | 1.316 | 0.017 | 0.012 | 52.0 | 59.0 | 1.07 |
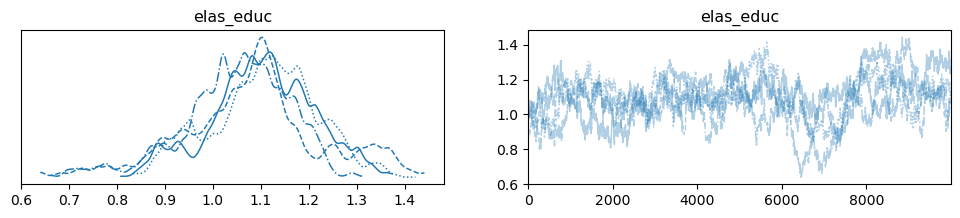
We can even see the marginal posterior for our elasticity estimate:
pm.plot_trace(samples, var_names = ['elas_educ']);
The predicted wage is included in the trace and is of dimension number of chains x number of samples x number of individuals. This is alot of information. To illustrate the information we have stored in our posterior let’s focus on only the first 50 individuals:
individs = [i for i in range(50)]
pm.plot_trace(samples.sel(wage_hat_dim_0 = individs), var_names=['wage_hat']);
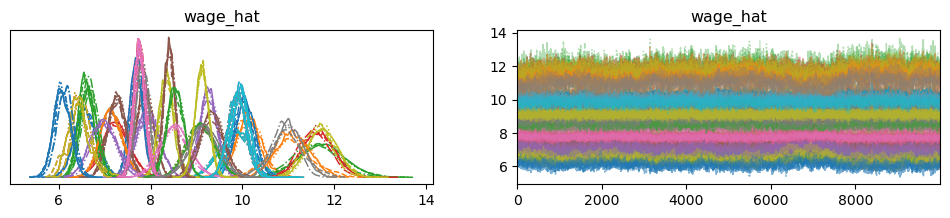
By storing these values in our trace, we can calculate policy relevant measures that immediately reflect the uncertainty of our parameter estimates since the distribution is being driven by the distribution of the posterior itself. This is evident in the posterior distributions of predicted wage for each individual.
Summary#
This workbook has demonstrated how to use pymc for a linear
regression model. We have examined some convergence criteria and see
that we have to increase sample size quite alot to meet the
Gelman-Rubin metric of r_hat \(\approx 1\). Even after increasing the
number of samples by quite alot, the values above fail to completely
satisfy this criteria for some of the parameters in our model.
In the next chapter we turn our attention to more modern step methods for achieving convergence with many fewer samples.